Organize pandas notebook with cool hacks (2020)
Does it ring a bell looking at this messy notebook? I am sure you must have created or encountered a similar kind of notebook while performing data analysis tasks in pandas.
Pandas is widely used by data scientists and ML Engineers all around the world to perform all kinds of data related tasks like data cleaning and preprocessing, data analysis, data manipulation, data conversion, etc. However, most of us are not using it right, as seen in the above example, which has decreased our productivity a lot.
You might wonder then what is the correct way to use pandas. Is there any particular way that we can make the notebook clean and modular so that we can increase our productivity?
Luckily, there is a type of quick hack or technique, whatever you may call it, which can be used to greatly improve the workflow and make notebooks not only clean and well organized but highly productive and efficient. The good thing is that you don’t need to install any extra packages or libraries. In the end, your notebook will look something like this.
Note: Dark mode is available on this website. You can switch between the modes by clicking the leftmost button at the bottom of the left sidebar.

Untitled12.ipynb
The way to achieve clean and well-organized pandas notebooks was explored in the presentation Untitled12.ipynb by Vincent D. Warmerdam at PyData Eindhoven 2019
The presentation Untitled12.ipynb: Prevent Miles of Scrolling, Reduce the Spaghetti Code from the Copy Pasta has been uploaded in youtube as well. You can watch the video below if you want:
In this article, I will briefly summarize the presentation by Vincent D. Warmerdam and then move on to the code implementation (solution) and a few code examples based on the methods used in his presentation.
The Untitled phenomena
He began his talk by introducing a term called Untitled phenomena. The term simply refers to the bad practice of not naming the notebook files which eventually creates an unorganized bunch of Untitled notebooks. As a result, he also named the presentation Untitled12.ipynb.
Moreover, not only the bad practice of naming that we follow but also the bad organization of code inside the notebook needs to be improved. Copying and pasting code multiple times creates spaghetti code. This is especially true for a lot of data science based Jupyter notebooks. The goal of his talk was to uncover a great pattern for pandas that would prevent loads of scrolling such that the code behaves like lego. He also gave some useful tricks and tips on how to prevent miles of scrolling and reduce the spaghetti code when creating Jupyter notebooks.
> Skip to coding solution
I have initially written a summary of the talk Untitled12.ipynb and explored some common problems in the usual coding style before moving to the solution. If you want to directly jump to the coding solution to create a clean pandas notebook using a pipeline, then click the link above. However, I recommend you to read the common problems I have mentioned before going to the solution.
Contents
I will be talking about the following topics which will more or less revolve around his talk.
- Importance of Workflow
- The Usual coding style
- Problems in the usual coding style
- Coding Solution
- Advantages
Importance of Workflow
At the beginning of the presentation, he began by discussing the following points that highlight the importance of workflows and the need of jupyter-notebook and pandas over excel:
We want to separate the data from the analysis: Tha analysis portion should not modify the raw data. The raw data should be safe from these modifications so that it can be reused later as well. However, this is not possible in excel.
We want to be able to automate our analysis. The main aim of programming and workflow is automation. Our tasks become a lot easier if we can automate the analysis using a pandas script rather than performing the analysis every time using Excel.
We want our analysis to be reproducible i.e. we must be able to reproduce the same analysis results on the data at a later time in the future.
We should not pay a third part obscene amounts of money for something as basic as arithmetic. This budget is better allocated towards innovation and education of staff.
However, the current style of coding in pandas and jupyter notebook has solved only the last point.
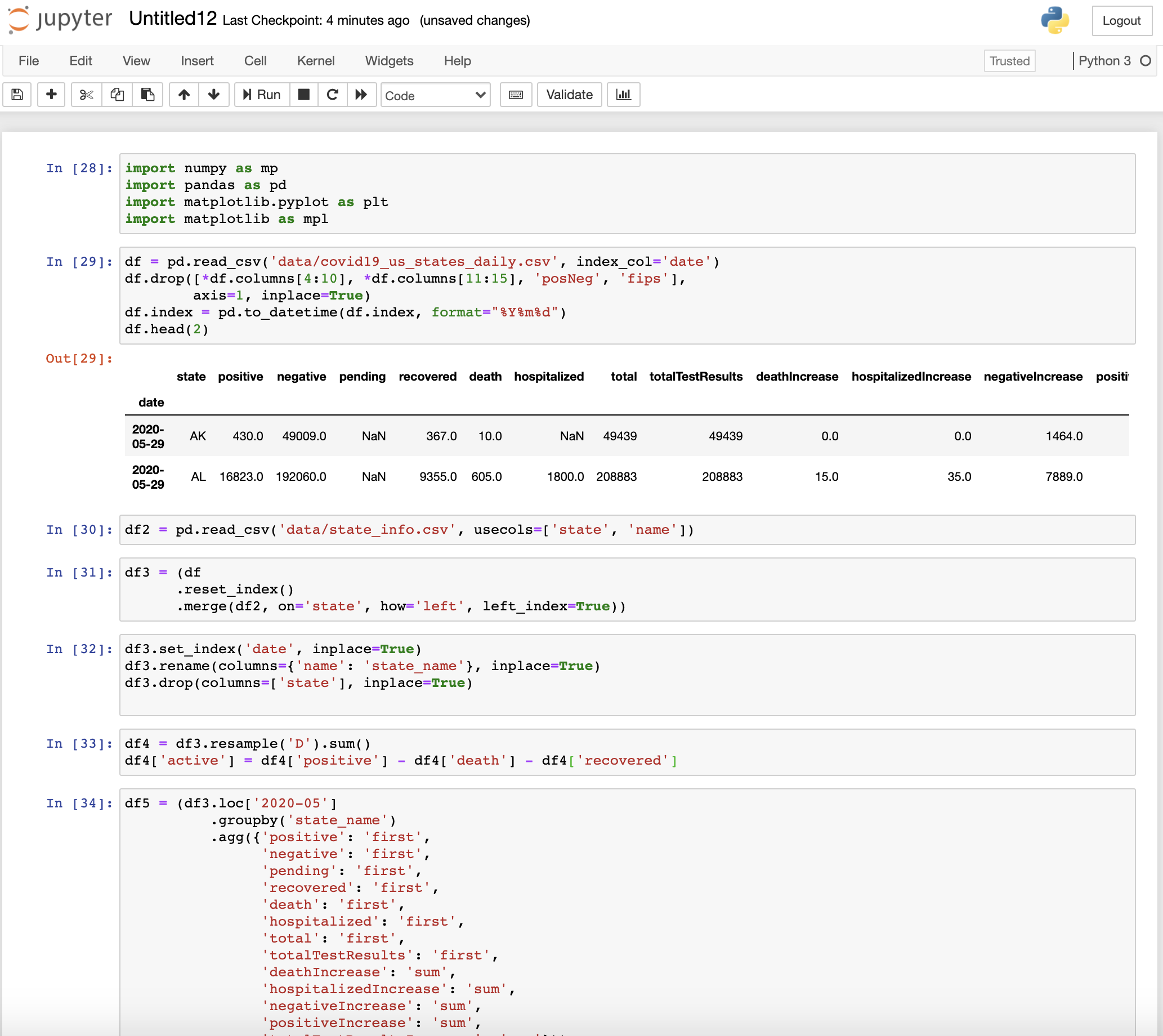
The usual coding style
Let’s explore the common practice of writing pandas code and try to point out the major problems in such approaches.
Initially, I will show the general workflow that most of us follow while using pandas. I will be performing some analysis on the real COVID 19 dataset of the U.S. states obtained from The COVID Tracking Project which is available under the Creative Commons CC BY-NC-4.0 license. The dataset is updated each day between 4 pm and 5 pm EDT.
After showing the common approach, I will point out the major pitfalls and then move on to the solution.
First, I will download the U.S. COVID-19 dataset using the API provided by The COVID Tracking Project
!mkdir data
!wget -O data/covid19_us_states_daily.csv https://covidtracking.com/api/v1/states/daily.csv
!wget -O data/state_info.csv https://covidtracking.com/api/v1/states/info.csv
--2020-06-05 16:34:10-- https://covidtracking.com/api/v1/states/daily.csv
Resolving covidtracking.com (covidtracking.com)... 104.248.63.231, 2604:a880:400:d1::888:7001
Connecting to covidtracking.com (covidtracking.com)|104.248.63.231|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: unspecified [text/csv]
Saving to: ‘data/covid19_us_states_daily.csv’
data/covid19_us_sta [ <=> ] 987.40K 3.11MB/s in 0.3s
2020-06-05 16:34:11 (3.11 MB/s) - ‘data/covid19_us_states_daily.csv’ saved [1011093]
--2020-06-05 16:34:12-- https://covidtracking.com/api/v1/states/info.csv
Resolving covidtracking.com (covidtracking.com)... 104.248.50.87, 2604:a880:400:d1::888:7001
Connecting to covidtracking.com (covidtracking.com)|104.248.50.87|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: unspecified [text/csv]
Saving to: ‘data/state_info.csv’
data/state_info.csv [ <=> ] 27.67K --.-KB/s in 0.02s
2020-06-05 16:34:13 (1.43 MB/s) - ‘data/state_info.csv’ saved [28329]
import pandas as pd
# Importing plotly library for plotting interactive graphs
import plotly.graph_objects as go
from plotly.subplots import make_subplots
import plotly.express as px
import chart_studio
import chart_studio.plotly as py
The first step is generally to read or import the data
df = pd.read_csv('data/covid19_us_states_daily.csv', index_col='date')
df.head()
| state | positive | negative | pending | hospitalizedCurrently | hospitalizedCumulative | inIcuCurrently | inIcuCumulative | onVentilatorCurrently | onVentilatorCumulative | recovered | dataQualityGrade | lastUpdateEt | dateModified | checkTimeEt | death | hospitalized | dateChecked | fips | positiveIncrease | negativeIncrease | total | totalTestResults | totalTestResultsIncrease | posNeg | deathIncrease | hospitalizedIncrease | hash | commercialScore | negativeRegularScore | negativeScore | positiveScore | score | grade | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| date | ||||||||||||||||||||||||||||||||||
| 20200604 | AK | 513.0 | 59584.0 | NaN | 13.0 | NaN | NaN | NaN | 1.0 | NaN | 376.0 | A | 6/4/2020 00:00 | 2020-06-04T00:00:00Z | 06/03 20:00 | 10.0 | NaN | 2020-06-04T00:00:00Z | 2 | 8 | 1907 | 60097 | 60097 | 1915 | 60097 | 0 | 0 | c1046011af7271cbe2e6698526714c6cb5b92748 | 0 | 0 | 0 | 0 | 0 | NaN |
| 20200604 | AL | 19072.0 | 216227.0 | NaN | NaN | 1929.0 | NaN | 601.0 | NaN | 357.0 | 11395.0 | B | 6/4/2020 00:00 | 2020-06-04T00:00:00Z | 06/03 20:00 | 653.0 | 1929.0 | 2020-06-04T00:00:00Z | 1 | 221 | 3484 | 235299 | 235299 | 3705 | 235299 | 0 | 29 | bcbefdb36212ba2b97b5a354f4e45bf16648ee23 | 0 | 0 | 0 | 0 | 0 | NaN |
| 20200604 | AR | 8067.0 | 134413.0 | NaN | 138.0 | 757.0 | NaN | NaN | 30.0 | 127.0 | 5717.0 | A | 6/4/2020 00:00 | 2020-06-04T00:00:00Z | 06/03 20:00 | 142.0 | 757.0 | 2020-06-04T00:00:00Z | 5 | 0 | 0 | 142480 | 142480 | 0 | 142480 | 0 | 26 | acd3a4fbbc3dbb32138725f91e3261d683e7052a | 0 | 0 | 0 | 0 | 0 | NaN |
| 20200604 | AS | 0.0 | 174.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | C | 6/1/2020 00:00 | 2020-06-01T00:00:00Z | 05/31 20:00 | 0.0 | NaN | 2020-06-01T00:00:00Z | 60 | 0 | 0 | 174 | 174 | 0 | 174 | 0 | 0 | 8bbc72fa42781e0549e2e4f9f4c3e7cbef14ab32 | 0 | 0 | 0 | 0 | 0 | NaN |
| 20200604 | AZ | 22753.0 | 227002.0 | NaN | 1079.0 | 3195.0 | 375.0 | NaN | 223.0 | NaN | 5172.0 | A+ | 6/4/2020 00:00 | 2020-06-04T00:00:00Z | 06/03 20:00 | 996.0 | 3195.0 | 2020-06-04T00:00:00Z | 4 | 520 | 4710 | 249755 | 249755 | 5230 | 249755 | 15 | 66 | 1fa237b8204cd23701577aef6338d339daa4452e | 0 | 0 | 0 | 0 | 0 | NaN |
After taking a glance at the data, I realize that the date is not formatted well, so I format it.
df.index = pd.to_datetime(df.index, format="%Y%m%d")
df.head()
| state | positive | negative | pending | hospitalizedCurrently | hospitalizedCumulative | inIcuCurrently | inIcuCumulative | onVentilatorCurrently | onVentilatorCumulative | recovered | dataQualityGrade | lastUpdateEt | dateModified | checkTimeEt | death | hospitalized | dateChecked | fips | positiveIncrease | negativeIncrease | total | totalTestResults | totalTestResultsIncrease | posNeg | deathIncrease | hospitalizedIncrease | hash | commercialScore | negativeRegularScore | negativeScore | positiveScore | score | grade | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| date | ||||||||||||||||||||||||||||||||||
| 2020-06-04 | AK | 513.0 | 59584.0 | NaN | 13.0 | NaN | NaN | NaN | 1.0 | NaN | 376.0 | A | 6/4/2020 00:00 | 2020-06-04T00:00:00Z | 06/03 20:00 | 10.0 | NaN | 2020-06-04T00:00:00Z | 2 | 8 | 1907 | 60097 | 60097 | 1915 | 60097 | 0 | 0 | c1046011af7271cbe2e6698526714c6cb5b92748 | 0 | 0 | 0 | 0 | 0 | NaN |
| 2020-06-04 | AL | 19072.0 | 216227.0 | NaN | NaN | 1929.0 | NaN | 601.0 | NaN | 357.0 | 11395.0 | B | 6/4/2020 00:00 | 2020-06-04T00:00:00Z | 06/03 20:00 | 653.0 | 1929.0 | 2020-06-04T00:00:00Z | 1 | 221 | 3484 | 235299 | 235299 | 3705 | 235299 | 0 | 29 | bcbefdb36212ba2b97b5a354f4e45bf16648ee23 | 0 | 0 | 0 | 0 | 0 | NaN |
| 2020-06-04 | AR | 8067.0 | 134413.0 | NaN | 138.0 | 757.0 | NaN | NaN | 30.0 | 127.0 | 5717.0 | A | 6/4/2020 00:00 | 2020-06-04T00:00:00Z | 06/03 20:00 | 142.0 | 757.0 | 2020-06-04T00:00:00Z | 5 | 0 | 0 | 142480 | 142480 | 0 | 142480 | 0 | 26 | acd3a4fbbc3dbb32138725f91e3261d683e7052a | 0 | 0 | 0 | 0 | 0 | NaN |
| 2020-06-04 | AS | 0.0 | 174.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | C | 6/1/2020 00:00 | 2020-06-01T00:00:00Z | 05/31 20:00 | 0.0 | NaN | 2020-06-01T00:00:00Z | 60 | 0 | 0 | 174 | 174 | 0 | 174 | 0 | 0 | 8bbc72fa42781e0549e2e4f9f4c3e7cbef14ab32 | 0 | 0 | 0 | 0 | 0 | NaN |
| 2020-06-04 | AZ | 22753.0 | 227002.0 | NaN | 1079.0 | 3195.0 | 375.0 | NaN | 223.0 | NaN | 5172.0 | A+ | 6/4/2020 00:00 | 2020-06-04T00:00:00Z | 06/03 20:00 | 996.0 | 3195.0 | 2020-06-04T00:00:00Z | 4 | 520 | 4710 | 249755 | 249755 | 5230 | 249755 | 15 | 66 | 1fa237b8204cd23701577aef6338d339daa4452e | 0 | 0 | 0 | 0 | 0 | NaN |
Then, I try to view some additional information about the dataset
df.info()
<class 'pandas.core.frame.DataFrame'>
DatetimeIndex: 5113 entries, 2020-06-04 to 2020-01-22
Data columns (total 34 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 state 5113 non-null object
1 positive 5098 non-null float64
2 negative 4902 non-null float64
3 pending 842 non-null float64
4 hospitalizedCurrently 2591 non-null float64
5 hospitalizedCumulative 2318 non-null float64
6 inIcuCurrently 1362 non-null float64
7 inIcuCumulative 576 non-null float64
8 onVentilatorCurrently 1157 non-null float64
9 onVentilatorCumulative 198 non-null float64
10 recovered 2409 non-null float64
11 dataQualityGrade 4012 non-null object
12 lastUpdateEt 4758 non-null object
13 dateModified 4758 non-null object
14 checkTimeEt 4758 non-null object
15 death 4388 non-null float64
16 hospitalized 2318 non-null float64
17 dateChecked 4758 non-null object
18 fips 5113 non-null int64
19 positiveIncrease 5113 non-null int64
20 negativeIncrease 5113 non-null int64
21 total 5113 non-null int64
22 totalTestResults 5113 non-null int64
23 totalTestResultsIncrease 5113 non-null int64
24 posNeg 5113 non-null int64
25 deathIncrease 5113 non-null int64
26 hospitalizedIncrease 5113 non-null int64
27 hash 5113 non-null object
28 commercialScore 5113 non-null int64
29 negativeRegularScore 5113 non-null int64
30 negativeScore 5113 non-null int64
31 positiveScore 5113 non-null int64
32 score 5113 non-null int64
33 grade 0 non-null float64
dtypes: float64(13), int64(14), object(7)
memory usage: 1.4+ MB
You can see that various columns are not of use. So, I decide to remove such columns.
df.drop([*df.columns[4:10], *df.columns[11:15], 'posNeg', 'fips'],
axis=1, inplace=True)
df.info()
<class 'pandas.core.frame.DataFrame'>
DatetimeIndex: 5113 entries, 2020-06-04 to 2020-01-22
Data columns (total 22 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 state 5113 non-null object
1 positive 5098 non-null float64
2 negative 4902 non-null float64
3 pending 842 non-null float64
4 recovered 2409 non-null float64
5 death 4388 non-null float64
6 hospitalized 2318 non-null float64
7 dateChecked 4758 non-null object
8 positiveIncrease 5113 non-null int64
9 negativeIncrease 5113 non-null int64
10 total 5113 non-null int64
11 totalTestResults 5113 non-null int64
12 totalTestResultsIncrease 5113 non-null int64
13 deathIncrease 5113 non-null int64
14 hospitalizedIncrease 5113 non-null int64
15 hash 5113 non-null object
16 commercialScore 5113 non-null int64
17 negativeRegularScore 5113 non-null int64
18 negativeScore 5113 non-null int64
19 positiveScore 5113 non-null int64
20 score 5113 non-null int64
21 grade 0 non-null float64
dtypes: float64(7), int64(12), object(3)
memory usage: 918.7+ KB
I also realize that there are a lot of missing (nan or null) values. So, I replace the missing values by 0.
df.fillna(value=0, inplace=True)
df.head()
| state | positive | negative | pending | recovered | death | hospitalized | dateChecked | positiveIncrease | negativeIncrease | total | totalTestResults | totalTestResultsIncrease | deathIncrease | hospitalizedIncrease | hash | commercialScore | negativeRegularScore | negativeScore | positiveScore | score | grade | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| date | ||||||||||||||||||||||
| 2020-06-04 | AK | 513.0 | 59584.0 | 0.0 | 376.0 | 10.0 | 0.0 | 2020-06-04T00:00:00Z | 8 | 1907 | 60097 | 60097 | 1915 | 0 | 0 | c1046011af7271cbe2e6698526714c6cb5b92748 | 0 | 0 | 0 | 0 | 0 | 0.0 |
| 2020-06-04 | AL | 19072.0 | 216227.0 | 0.0 | 11395.0 | 653.0 | 1929.0 | 2020-06-04T00:00:00Z | 221 | 3484 | 235299 | 235299 | 3705 | 0 | 29 | bcbefdb36212ba2b97b5a354f4e45bf16648ee23 | 0 | 0 | 0 | 0 | 0 | 0.0 |
| 2020-06-04 | AR | 8067.0 | 134413.0 | 0.0 | 5717.0 | 142.0 | 757.0 | 2020-06-04T00:00:00Z | 0 | 0 | 142480 | 142480 | 0 | 0 | 26 | acd3a4fbbc3dbb32138725f91e3261d683e7052a | 0 | 0 | 0 | 0 | 0 | 0.0 |
| 2020-06-04 | AS | 0.0 | 174.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2020-06-01T00:00:00Z | 0 | 0 | 174 | 174 | 0 | 0 | 0 | 8bbc72fa42781e0549e2e4f9f4c3e7cbef14ab32 | 0 | 0 | 0 | 0 | 0 | 0.0 |
| 2020-06-04 | AZ | 22753.0 | 227002.0 | 0.0 | 5172.0 | 996.0 | 3195.0 | 2020-06-04T00:00:00Z | 520 | 4710 | 249755 | 249755 | 5230 | 15 | 66 | 1fa237b8204cd23701577aef6338d339daa4452e | 0 | 0 | 0 | 0 | 0 | 0.0 |
I also want to add a column corresponding to the state name instead of the abbreviation. So, I merge state_info with the current dataframe.
df2 = pd.read_csv('data/state_info.csv', usecols=['state', 'name'])
df3 = (df
.reset_index()
.merge(df2, on='state', how='left', left_index=True))
df3.head()
| date | state | positive | negative | pending | recovered | death | hospitalized | dateChecked | positiveIncrease | negativeIncrease | total | totalTestResults | totalTestResultsIncrease | deathIncrease | hospitalizedIncrease | hash | commercialScore | negativeRegularScore | negativeScore | positiveScore | score | grade | name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2020-06-04 | AK | 513.0 | 59584.0 | 0.0 | 376.0 | 10.0 | 0.0 | 2020-06-04T00:00:00Z | 8 | 1907 | 60097 | 60097 | 1915 | 0 | 0 | c1046011af7271cbe2e6698526714c6cb5b92748 | 0 | 0 | 0 | 0 | 0 | 0.0 | Alaska |
| 1 | 2020-06-04 | AL | 19072.0 | 216227.0 | 0.0 | 11395.0 | 653.0 | 1929.0 | 2020-06-04T00:00:00Z | 221 | 3484 | 235299 | 235299 | 3705 | 0 | 29 | bcbefdb36212ba2b97b5a354f4e45bf16648ee23 | 0 | 0 | 0 | 0 | 0 | 0.0 | Alabama |
| 2 | 2020-06-04 | AR | 8067.0 | 134413.0 | 0.0 | 5717.0 | 142.0 | 757.0 | 2020-06-04T00:00:00Z | 0 | 0 | 142480 | 142480 | 0 | 0 | 26 | acd3a4fbbc3dbb32138725f91e3261d683e7052a | 0 | 0 | 0 | 0 | 0 | 0.0 | Arkansas |
| 3 | 2020-06-04 | AS | 0.0 | 174.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2020-06-01T00:00:00Z | 0 | 0 | 174 | 174 | 0 | 0 | 0 | 8bbc72fa42781e0549e2e4f9f4c3e7cbef14ab32 | 0 | 0 | 0 | 0 | 0 | 0.0 | American Samoa |
| 4 | 2020-06-04 | AZ | 22753.0 | 227002.0 | 0.0 | 5172.0 | 996.0 | 3195.0 | 2020-06-04T00:00:00Z | 520 | 4710 | 249755 | 249755 | 5230 | 15 | 66 | 1fa237b8204cd23701577aef6338d339daa4452e | 0 | 0 | 0 | 0 | 0 | 0.0 | Arizona |
I realize that the date index is lost. So, I reset the date index. Also, it is better to rename the column name as state_name.
df3.set_index('date', inplace=True)
df3.rename(columns={'name': 'state_name'}, inplace=True)
df3.head()
| state | positive | negative | pending | recovered | death | hospitalized | dateChecked | positiveIncrease | negativeIncrease | total | totalTestResults | totalTestResultsIncrease | deathIncrease | hospitalizedIncrease | hash | commercialScore | negativeRegularScore | negativeScore | positiveScore | score | grade | state_name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| date | |||||||||||||||||||||||
| 2020-06-04 | AK | 513.0 | 59584.0 | 0.0 | 376.0 | 10.0 | 0.0 | 2020-06-04T00:00:00Z | 8 | 1907 | 60097 | 60097 | 1915 | 0 | 0 | c1046011af7271cbe2e6698526714c6cb5b92748 | 0 | 0 | 0 | 0 | 0 | 0.0 | Alaska |
| 2020-06-04 | AL | 19072.0 | 216227.0 | 0.0 | 11395.0 | 653.0 | 1929.0 | 2020-06-04T00:00:00Z | 221 | 3484 | 235299 | 235299 | 3705 | 0 | 29 | bcbefdb36212ba2b97b5a354f4e45bf16648ee23 | 0 | 0 | 0 | 0 | 0 | 0.0 | Alabama |
| 2020-06-04 | AR | 8067.0 | 134413.0 | 0.0 | 5717.0 | 142.0 | 757.0 | 2020-06-04T00:00:00Z | 0 | 0 | 142480 | 142480 | 0 | 0 | 26 | acd3a4fbbc3dbb32138725f91e3261d683e7052a | 0 | 0 | 0 | 0 | 0 | 0.0 | Arkansas |
| 2020-06-04 | AS | 0.0 | 174.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2020-06-01T00:00:00Z | 0 | 0 | 174 | 174 | 0 | 0 | 0 | 8bbc72fa42781e0549e2e4f9f4c3e7cbef14ab32 | 0 | 0 | 0 | 0 | 0 | 0.0 | American Samoa |
| 2020-06-04 | AZ | 22753.0 | 227002.0 | 0.0 | 5172.0 | 996.0 | 3195.0 | 2020-06-04T00:00:00Z | 520 | 4710 | 249755 | 249755 | 5230 | 15 | 66 | 1fa237b8204cd23701577aef6338d339daa4452e | 0 | 0 | 0 | 0 | 0 | 0.0 | Arizona |
Now that the data is ready for some analysis, I decide to plot deaths count in each state indexed by date using the interactive plotly library.
fig1 = px.line(df3, x=df3.index, y='death', color='state')
fig1.update_layout(xaxis_title='date', title='Total deaths in each state (Cumulative)')
py.plot(fig1, filename = 'daily_deaths', auto_open=True)
'https://plotly.com/~ayush.kumar.shah/1/'
Note: These plots are interactive, so you can zoom in or out, pinch, hover over the graph, download it, and so on.
Now, I decide to calculate the total deaths in the US across all states and plot it.
df4 = df3.resample('D').sum()
df4.head()
| positive | negative | pending | recovered | death | hospitalized | positiveIncrease | negativeIncrease | total | totalTestResults | totalTestResultsIncrease | deathIncrease | hospitalizedIncrease | commercialScore | negativeRegularScore | negativeScore | positiveScore | score | grade | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| date | |||||||||||||||||||
| 2020-01-22 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 |
| 2020-01-23 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 |
| 2020-01-24 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 |
| 2020-01-25 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 |
| 2020-01-26 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 |
fig2 = px.line(df4, x=df4.index, y='death')
fig2.update_layout(xaxis_title='date', title='Total deaths in the U.S. (Cumulative)')
py.plot(fig2, filename = 'total_daily_deaths', auto_open=True)
'https://plotly.com/~ayush.kumar.shah/4/'
I also want to calculate the number of Active cases i.e.
Active = positive - deaths - recovered
df4['active'] = df4['positive'] - df4['death'] - df4['recovered']
Now, after calculating the active column, I want to plot active cases instead of death. So, I go to the previous cell and replace death by active and generate the plot.
In [25]: df4[’death’].plot()
In [25]: df4[‘active’].plot()
fig3 = px.line(df4, x=df4.index, y='active')
fig3.update_layout(xaxis_title='date', title='Total active cases in the U.S. (Cumulative)')
py.plot(fig3, filename = 'total_daily_active', auto_open=True)
'https://plotly.com/~ayush.kumar.shah/6/'
Then I decide to calculate the statistics of a single month of May only. Since the data is cumulative, I need to subtract the data of May from data of April to find the increase in various statistics in May after which I plot the results.
df5 = (df3.loc['2020-05']
.groupby('state_name')
.agg({'positive': 'first',
'negative': 'first',
'pending': 'first',
'recovered': 'first',
'death': 'first',
'hospitalized': 'first',
'total': 'first',
'totalTestResults': 'first',
'deathIncrease': 'sum',
'hospitalizedIncrease': 'sum',
'negativeIncrease': 'sum',
'positiveIncrease': 'sum',
'totalTestResultsIncrease': 'sum'}))
df6 = (df3.loc['2020-04']
.groupby('state_name')
.agg({'positive': 'first',
'negative': 'first',
'pending': 'first',
'recovered': 'first',
'death': 'first',
'hospitalized': 'first',
'total': 'first',
'totalTestResults': 'first',
'deathIncrease': 'sum',
'hospitalizedIncrease': 'sum',
'negativeIncrease': 'sum',
'positiveIncrease': 'sum',
'totalTestResultsIncrease': 'sum'}))
df7 = df5.sub(df6)
df7.head()
| positive | negative | pending | recovered | death | hospitalized | total | totalTestResults | deathIncrease | hospitalizedIncrease | negativeIncrease | positiveIncrease | totalTestResultsIncrease | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| state_name | |||||||||||||
| Alabama | 10884.0 | 119473.0 | 0.0 | 9355.0 | 362.0 | 866.0 | 130357 | 130357 | 106 | -112 | 45594 | 4846 | 50440 |
| Alaska | 79.0 | 32497.0 | 0.0 | 116.0 | 1.0 | 0.0 | 32576 | 32576 | -5 | 7 | 17327 | -157 | 17170 |
| American Samoa | 0.0 | 171.0 | -17.0 | 0.0 | 0.0 | 0.0 | 154 | 171 | 0 | 0 | 171 | 0 | 171 |
| Arizona | 12288.0 | 141132.0 | 0.0 | 3262.0 | 586.0 | 1829.0 | 153420 | 153420 | 290 | 660 | 95076 | 5929 | 101005 |
| Arkansas | 3998.0 | 77138.0 | 0.0 | 3970.0 | 72.0 | 309.0 | 81136 | 81136 | 19 | -93 | 37973 | 1266 | 39239 |
fig4 = px.bar(df7, x=df7.index, y='death')
fig4.update_layout(xaxis_title='state_name', title='Total Deaths in th US in May only')
py.plot(fig4, filename = 'total_deaths_May', auto_open=True)
'https://plotly.com/~ayush.kumar.shah/12/'
Problems in the usual coding style
Now that I have demonstrated the usual approach followed in pandas notebook, let’s discuss the problems in this approach.
1. Flow is disrupted:
The flow of the notebook is very difficult to understand and also creates problems. For example, we may create a variable name under the plot that needs it. In the above code as well, we created df3['active'] below the cell in which it is needed. So, it may cause errors when run by others. Also, you may have to scroll the notebook for miles and miles.
2. No reproducibility:
When the notebook is shared with others, the other person faces a lot of problems to execute or understand the notebook. For instance, the name of the dataframes doesn’t signify any information about the type of dataframe. It runs from df1 to df7 and creates a lot of confusion. But you want to create a notebook which is very easy to iterate on and the one you can share with your colleagues.
3. Difficult to move the code to production:
With this approach, your code is not ready to move into production. You end up having to rewrite the whole notebook before moving it to production which is not effective.
4. Unable to automate:
The notebook in the current condition cannot be automated for analysis since there may occur a lot of problems like an error in code execution, unavailability of filenames used.
Although the code may give an interesting conclusion or desired output, we are not quite sure that conclusion is at least correct.
Despite having so many problems associated with this approach, it is common for everyone to still use this type of flow while making a notebook since while coding, people enjoy when the code works when they check the outputs and hence keep on similarly continuing the coding.
Coding Solution
1. Naming convention
Follow a naming convention for the notebook according to the task as suggested by Cookiecutter Data Science that shows the owner and the order the analysis was done in. You can use the format
<step>-<ghuser>-<description>.ipynb
(e.g., 0.1-ayush-visualize-corona-us.ipynb).
2. Plan your steps beforehand
Load the data and then think in advance about all the steps of analysis or tasks you could be doing in the notebook. You don’t need to think the logic right away but just keep in mind the steps.
df = pd.read_csv('data/covid19_us_states_daily.csv', index_col='date')
3. Create functions
You know that initially, you want to clean the data and make sure the columns and indexes are in a proper usable format. So, why not create a function for that and name it according to the subtasks on the dataframe.
For example, initially you want to make the index a proper datetime object. Then you may want to do remove the duplicates, then add state name. Just add these functions without even thinking the logic and then later you can add the logic. This way, you will be on track and not lost.
The functions are created after creating the decorator.
4. Create proper decorators
Before adding functions, let’s also think about some additional utility that would be helpful. During the pandas analysis, you often check the shape, columns, and other information associated to the dataframe after performing an operation. However, a decorator can help automate this process.
Decorator is simply a function that expects a function and returns a function. It’s really functional right, haha. Don’t get confused by the definition. It is not so difficult as it sounds. We will see how it works in the code below.
Also, if you are not familiar with decorators or want to learn more about it, you can visit the article by Geir Arne Hjelle.
import datetime as dt
def df_info(f):
def wrapper(df, *args, **kwargs):
tic = dt.datetime.now()
result = f(df, *args, **kwargs)
toc = dt.datetime.now()
print("\n\n{} took {} time\n".format(f.__name__, toc - tic))
print("After applying {}\n".format(f.__name__))
print("Shape of df = {}\n".format(result.shape))
print("Columns of df are {}\n".format(result.columns))
print("Index of df is {}\n".format(result.index))
for i in range(100): print("-", end='')
return result
return wrapper
We have created a decorator called df_info which displays information like time taken by the function, shape, and columns after applying any function f.
The advantage of using a deorator is that we get logging. You can modify the decorator according to the information that you want to log or display after performing an operation on the dataframe.
Now, we create functions as our plan and use these decorators on them by using @df_info. This will be equivalent to calling df_info(f(df, *args, **kwargs))
@df_info
def create_dateindex(df):
df.index = pd.to_datetime(df.index, format="%Y%m%d")
return df
@df_info
def remove_columns(df):
df.drop([*df.columns[4:10], *df.columns[11:15], 'posNeg', 'fips'],
axis=1, inplace=True)
return df
@df_info
def fill_missing(df):
df.fillna(value=0, inplace=True)
return df
@df_info
def add_state_name(df):
_df = pd.read_csv('data/state_info.csv', usecols=['state', 'name'])
df = (df
.reset_index()
.merge(_df, on='state', how='left', left_index=True))
df.set_index('date', inplace=True)
df.rename(columns={'name': 'state_name'}, inplace=True)
return df
@df_info
def drop_state(df):
df.drop(columns=['state'], inplace=True)
return df
@df_info
def sample_daily(df):
df = df.resample('D').sum()
return df
@df_info
def add_active_cases(df):
df['active'] = df['positive'] - df['death'] - df['recovered']
return df
def aggregate_monthly(df, month):
df = (df.loc[month]
.groupby('state_name')
.agg({'positive': 'first',
'negative': 'first',
'pending': 'first',
'recovered': 'first',
'death': 'first',
'hospitalized': 'first',
'total': 'first',
'totalTestResults': 'first',
'deathIncrease': 'sum',
'hospitalizedIncrease': 'sum',
'negativeIncrease': 'sum',
'positiveIncrease': 'sum',
'totalTestResultsIncrease': 'sum'}))
return df
@df_info
def create_month_only(df, month):
df_current = aggregate_monthly(df, month)
if int(month[-2:]) == 0:
prev_month = str(int(month[:4]) - 1) + '-12'
else:
prev_month = month[:5] + '{:02d}'.format(int(month[-2:])-1)
df_previous = aggregate_monthly(df, prev_month)
df = df_current.sub(df_previous)
return df
5. Remove side effect
However, these functions make changes that are inplace (side effects) i.e. modifies the originally loaded dataframe. So, to solve this, we add a function called start pipeline, which returns a copy of dataframe.
def start_pipeline(df):
return df.copy()
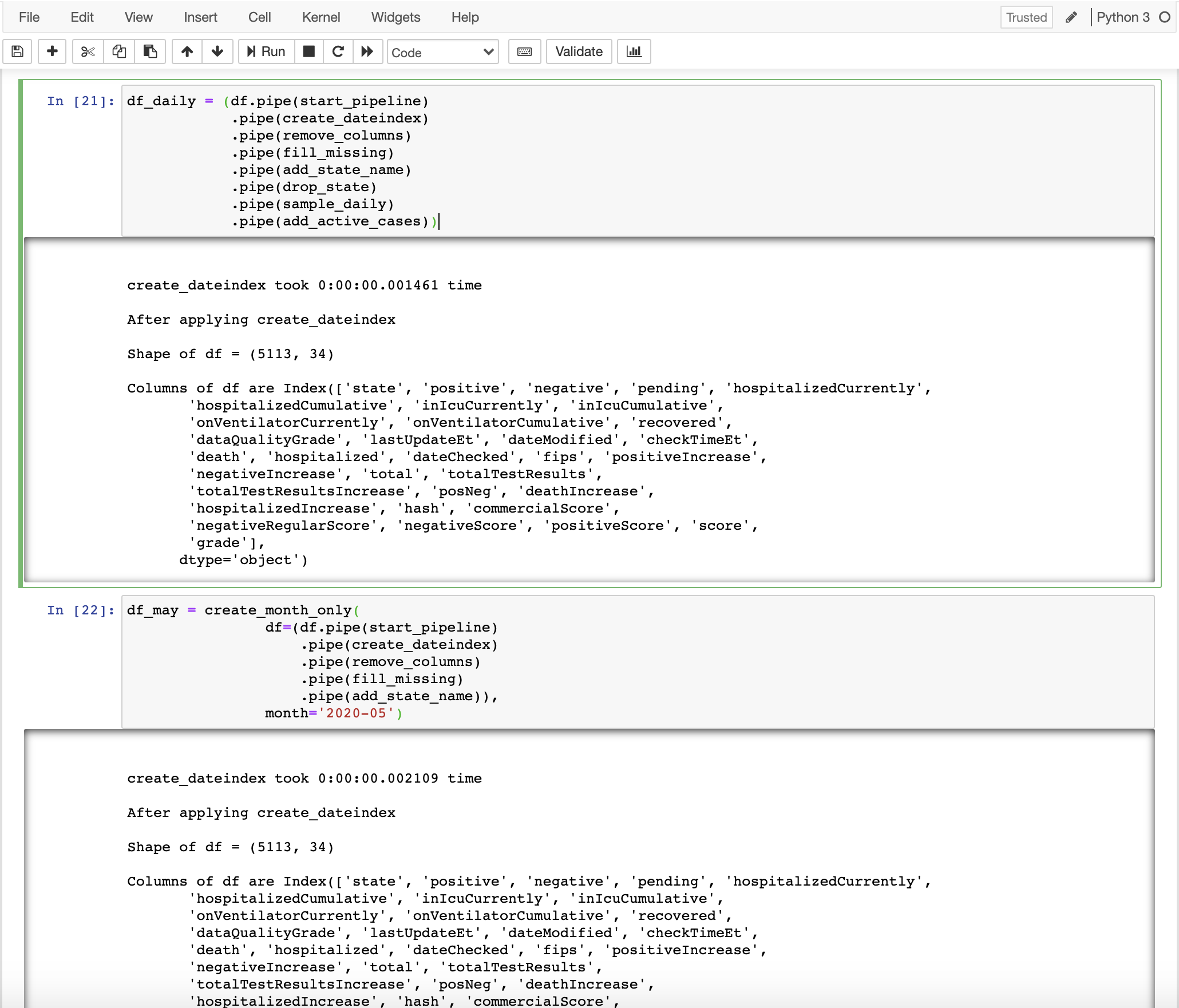
6. Constructing pandas pipelines (Main step)
Now, let’s use these functions to achieve the previous tasks using pipe
df_daily = (df.pipe(start_pipeline)
.pipe(create_dateindex)
.pipe(remove_columns)
.pipe(fill_missing)
.pipe(add_state_name)
.pipe(sample_daily)
.pipe(add_active_cases))
create_dateindex took 0:00:00.003388 time
After applying create_dateindex
Shape of df = (5113, 34)
Columns of df are Index(['state', 'positive', 'negative', 'pending', 'hospitalizedCurrently',
'hospitalizedCumulative', 'inIcuCurrently', 'inIcuCumulative',
'onVentilatorCurrently', 'onVentilatorCumulative', 'recovered',
'dataQualityGrade', 'lastUpdateEt', 'dateModified', 'checkTimeEt',
'death', 'hospitalized', 'dateChecked', 'fips', 'positiveIncrease',
'negativeIncrease', 'total', 'totalTestResults',
'totalTestResultsIncrease', 'posNeg', 'deathIncrease',
'hospitalizedIncrease', 'hash', 'commercialScore',
'negativeRegularScore', 'negativeScore', 'positiveScore', 'score',
'grade'],
dtype='object')
Index of df is DatetimeIndex(['2020-06-04', '2020-06-04', '2020-06-04', '2020-06-04',
'2020-06-04', '2020-06-04', '2020-06-04', '2020-06-04',
'2020-06-04', '2020-06-04',
...
'2020-01-31', '2020-01-30', '2020-01-29', '2020-01-28',
'2020-01-27', '2020-01-26', '2020-01-25', '2020-01-24',
'2020-01-23', '2020-01-22'],
dtype='datetime64[ns]', name='date', length=5113, freq=None)
----------------------------------------------------------------------------------------------------
remove_columns took 0:00:00.002087 time
After applying remove_columns
Shape of df = (5113, 22)
Columns of df are Index(['state', 'positive', 'negative', 'pending', 'recovered', 'death',
'hospitalized', 'dateChecked', 'positiveIncrease', 'negativeIncrease',
'total', 'totalTestResults', 'totalTestResultsIncrease',
'deathIncrease', 'hospitalizedIncrease', 'hash', 'commercialScore',
'negativeRegularScore', 'negativeScore', 'positiveScore', 'score',
'grade'],
dtype='object')
Index of df is DatetimeIndex(['2020-06-04', '2020-06-04', '2020-06-04', '2020-06-04',
'2020-06-04', '2020-06-04', '2020-06-04', '2020-06-04',
'2020-06-04', '2020-06-04',
...
'2020-01-31', '2020-01-30', '2020-01-29', '2020-01-28',
'2020-01-27', '2020-01-26', '2020-01-25', '2020-01-24',
'2020-01-23', '2020-01-22'],
dtype='datetime64[ns]', name='date', length=5113, freq=None)
----------------------------------------------------------------------------------------------------
fill_missing took 0:00:00.006381 time
After applying fill_missing
Shape of df = (5113, 22)
Columns of df are Index(['state', 'positive', 'negative', 'pending', 'recovered', 'death',
'hospitalized', 'dateChecked', 'positiveIncrease', 'negativeIncrease',
'total', 'totalTestResults', 'totalTestResultsIncrease',
'deathIncrease', 'hospitalizedIncrease', 'hash', 'commercialScore',
'negativeRegularScore', 'negativeScore', 'positiveScore', 'score',
'grade'],
dtype='object')
Index of df is DatetimeIndex(['2020-06-04', '2020-06-04', '2020-06-04', '2020-06-04',
'2020-06-04', '2020-06-04', '2020-06-04', '2020-06-04',
'2020-06-04', '2020-06-04',
...
'2020-01-31', '2020-01-30', '2020-01-29', '2020-01-28',
'2020-01-27', '2020-01-26', '2020-01-25', '2020-01-24',
'2020-01-23', '2020-01-22'],
dtype='datetime64[ns]', name='date', length=5113, freq=None)
----------------------------------------------------------------------------------------------------
add_state_name took 0:00:00.015122 time
After applying add_state_name
Shape of df = (5113, 23)
Columns of df are Index(['state', 'positive', 'negative', 'pending', 'recovered', 'death',
'hospitalized', 'dateChecked', 'positiveIncrease', 'negativeIncrease',
'total', 'totalTestResults', 'totalTestResultsIncrease',
'deathIncrease', 'hospitalizedIncrease', 'hash', 'commercialScore',
'negativeRegularScore', 'negativeScore', 'positiveScore', 'score',
'grade', 'state_name'],
dtype='object')
Index of df is DatetimeIndex(['2020-06-04', '2020-06-04', '2020-06-04', '2020-06-04',
'2020-06-04', '2020-06-04', '2020-06-04', '2020-06-04',
'2020-06-04', '2020-06-04',
...
'2020-01-31', '2020-01-30', '2020-01-29', '2020-01-28',
'2020-01-27', '2020-01-26', '2020-01-25', '2020-01-24',
'2020-01-23', '2020-01-22'],
dtype='datetime64[ns]', name='date', length=5113, freq=None)
----------------------------------------------------------------------------------------------------
sample_daily took 0:00:00.017170 time
After applying sample_daily
Shape of df = (135, 19)
Columns of df are Index(['positive', 'negative', 'pending', 'recovered', 'death', 'hospitalized',
'positiveIncrease', 'negativeIncrease', 'total', 'totalTestResults',
'totalTestResultsIncrease', 'deathIncrease', 'hospitalizedIncrease',
'commercialScore', 'negativeRegularScore', 'negativeScore',
'positiveScore', 'score', 'grade'],
dtype='object')
Index of df is DatetimeIndex(['2020-01-22', '2020-01-23', '2020-01-24', '2020-01-25',
'2020-01-26', '2020-01-27', '2020-01-28', '2020-01-29',
'2020-01-30', '2020-01-31',
...
'2020-05-26', '2020-05-27', '2020-05-28', '2020-05-29',
'2020-05-30', '2020-05-31', '2020-06-01', '2020-06-02',
'2020-06-03', '2020-06-04'],
dtype='datetime64[ns]', name='date', length=135, freq='D')
----------------------------------------------------------------------------------------------------
add_active_cases took 0:00:00.002020 time
After applying add_active_cases
Shape of df = (135, 20)
Columns of df are Index(['positive', 'negative', 'pending', 'recovered', 'death', 'hospitalized',
'positiveIncrease', 'negativeIncrease', 'total', 'totalTestResults',
'totalTestResultsIncrease', 'deathIncrease', 'hospitalizedIncrease',
'commercialScore', 'negativeRegularScore', 'negativeScore',
'positiveScore', 'score', 'grade', 'active'],
dtype='object')
Index of df is DatetimeIndex(['2020-01-22', '2020-01-23', '2020-01-24', '2020-01-25',
'2020-01-26', '2020-01-27', '2020-01-28', '2020-01-29',
'2020-01-30', '2020-01-31',
...
'2020-05-26', '2020-05-27', '2020-05-28', '2020-05-29',
'2020-05-30', '2020-05-31', '2020-06-01', '2020-06-02',
'2020-06-03', '2020-06-04'],
dtype='datetime64[ns]', name='date', length=135, freq='D')
Check out all the logs displayed above. We are able to view in detail how each operation changed the data without having to print the dataframe after each operation.
fig2 = px.line(df_daily, x=df_daily.index, y='death')
fig2.update_layout(xaxis_title='date', title='Total deaths in the U.S. (Cumulative)')
py.plot(fig2, filename = 'total_daily_deaths', auto_open=True)
'https://plotly.com/~ayush.kumar.shah/4/'
fig3 = px.line(df_daily, x=df_daily.index, y='active')
fig3.update_layout(xaxis_title='date', title='Total active cases in the U.S. (Cumulative)')
py.plot(fig3, filename = 'total_daily_active', auto_open=True)
'https://plotly.com/~ayush.kumar.shah/6/'
df_may = create_month_only(
df=(df.pipe(start_pipeline)
.pipe(create_dateindex)
.pipe(remove_columns)
.pipe(fill_missing)
.pipe(add_state_name)),
month='2020-05')
create_dateindex took 0:00:00.002492 time
After applying create_dateindex
Shape of df = (5113, 34)
Columns of df are Index(['state', 'positive', 'negative', 'pending', 'hospitalizedCurrently',
'hospitalizedCumulative', 'inIcuCurrently', 'inIcuCumulative',
'onVentilatorCurrently', 'onVentilatorCumulative', 'recovered',
'dataQualityGrade', 'lastUpdateEt', 'dateModified', 'checkTimeEt',
'death', 'hospitalized', 'dateChecked', 'fips', 'positiveIncrease',
'negativeIncrease', 'total', 'totalTestResults',
'totalTestResultsIncrease', 'posNeg', 'deathIncrease',
'hospitalizedIncrease', 'hash', 'commercialScore',
'negativeRegularScore', 'negativeScore', 'positiveScore', 'score',
'grade'],
dtype='object')
Index of df is DatetimeIndex(['2020-06-04', '2020-06-04', '2020-06-04', '2020-06-04',
'2020-06-04', '2020-06-04', '2020-06-04', '2020-06-04',
'2020-06-04', '2020-06-04',
...
'2020-01-31', '2020-01-30', '2020-01-29', '2020-01-28',
'2020-01-27', '2020-01-26', '2020-01-25', '2020-01-24',
'2020-01-23', '2020-01-22'],
dtype='datetime64[ns]', name='date', length=5113, freq=None)
----------------------------------------------------------------------------------------------------
remove_columns took 0:00:00.002219 time
After applying remove_columns
Shape of df = (5113, 22)
Columns of df are Index(['state', 'positive', 'negative', 'pending', 'recovered', 'death',
'hospitalized', 'dateChecked', 'positiveIncrease', 'negativeIncrease',
'total', 'totalTestResults', 'totalTestResultsIncrease',
'deathIncrease', 'hospitalizedIncrease', 'hash', 'commercialScore',
'negativeRegularScore', 'negativeScore', 'positiveScore', 'score',
'grade'],
dtype='object')
Index of df is DatetimeIndex(['2020-06-04', '2020-06-04', '2020-06-04', '2020-06-04',
'2020-06-04', '2020-06-04', '2020-06-04', '2020-06-04',
'2020-06-04', '2020-06-04',
...
'2020-01-31', '2020-01-30', '2020-01-29', '2020-01-28',
'2020-01-27', '2020-01-26', '2020-01-25', '2020-01-24',
'2020-01-23', '2020-01-22'],
dtype='datetime64[ns]', name='date', length=5113, freq=None)
----------------------------------------------------------------------------------------------------
fill_missing took 0:00:00.001883 time
After applying fill_missing
Shape of df = (5113, 22)
Columns of df are Index(['state', 'positive', 'negative', 'pending', 'recovered', 'death',
'hospitalized', 'dateChecked', 'positiveIncrease', 'negativeIncrease',
'total', 'totalTestResults', 'totalTestResultsIncrease',
'deathIncrease', 'hospitalizedIncrease', 'hash', 'commercialScore',
'negativeRegularScore', 'negativeScore', 'positiveScore', 'score',
'grade'],
dtype='object')
Index of df is DatetimeIndex(['2020-06-04', '2020-06-04', '2020-06-04', '2020-06-04',
'2020-06-04', '2020-06-04', '2020-06-04', '2020-06-04',
'2020-06-04', '2020-06-04',
...
'2020-01-31', '2020-01-30', '2020-01-29', '2020-01-28',
'2020-01-27', '2020-01-26', '2020-01-25', '2020-01-24',
'2020-01-23', '2020-01-22'],
dtype='datetime64[ns]', name='date', length=5113, freq=None)
----------------------------------------------------------------------------------------------------
add_state_name took 0:00:00.014981 time
After applying add_state_name
Shape of df = (5113, 23)
Columns of df are Index(['state', 'positive', 'negative', 'pending', 'recovered', 'death',
'hospitalized', 'dateChecked', 'positiveIncrease', 'negativeIncrease',
'total', 'totalTestResults', 'totalTestResultsIncrease',
'deathIncrease', 'hospitalizedIncrease', 'hash', 'commercialScore',
'negativeRegularScore', 'negativeScore', 'positiveScore', 'score',
'grade', 'state_name'],
dtype='object')
Index of df is DatetimeIndex(['2020-06-04', '2020-06-04', '2020-06-04', '2020-06-04',
'2020-06-04', '2020-06-04', '2020-06-04', '2020-06-04',
'2020-06-04', '2020-06-04',
...
'2020-01-31', '2020-01-30', '2020-01-29', '2020-01-28',
'2020-01-27', '2020-01-26', '2020-01-25', '2020-01-24',
'2020-01-23', '2020-01-22'],
dtype='datetime64[ns]', name='date', length=5113, freq=None)
----------------------------------------------------------------------------------------------------
create_month_only took 0:00:00.031071 time
After applying create_month_only
Shape of df = (56, 13)
Columns of df are Index(['positive', 'negative', 'pending', 'recovered', 'death', 'hospitalized',
'total', 'totalTestResults', 'deathIncrease', 'hospitalizedIncrease',
'negativeIncrease', 'positiveIncrease', 'totalTestResultsIncrease'],
dtype='object')
Index of df is Index(['Alabama', 'Alaska', 'American Samoa', 'Arizona', 'Arkansas',
'California', 'Colorado', 'Connecticut', 'Delaware',
'District Of Columbia', 'Florida', 'Georgia', 'Guam', 'Hawaii', 'Idaho',
'Illinois', 'Indiana', 'Iowa', 'Kansas', 'Kentucky', 'Louisiana',
'Maine', 'Maryland', 'Massachusetts', 'Michigan', 'Minnesota',
'Mississippi', 'Missouri', 'Montana', 'Nebraska', 'Nevada',
'New Hampshire', 'New Jersey', 'New Mexico', 'New York',
'North Carolina', 'North Dakota', 'Northern Mariana Islands', 'Ohio',
'Oklahoma', 'Oregon', 'Pennsylvania', 'Puerto Rico', 'Rhode Island',
'South Carolina', 'South Dakota', 'Tennessee', 'Texas',
'US Virgin Islands', 'Utah', 'Vermont', 'Virginia', 'Washington',
'West Virginia', 'Wisconsin', 'Wyoming'],
dtype='object', name='state_name')
fig4 = px.bar(df_may, x=df_may.index, y='death')
fig4.update_layout(xaxis_title='state_name', title='Total Deaths in th US in May only')
py.plot(fig4, filename = 'total_deaths_May', auto_open=True)
'https://plotly.com/~ayush.kumar.shah/12/'
You can observe how easily pipe functionality has achieved the required task in a clean and organized way. Also, the original dataframe is intact and not affected by the above operations.
df.head()
| state | positive | negative | pending | hospitalizedCurrently | hospitalizedCumulative | inIcuCurrently | inIcuCumulative | onVentilatorCurrently | onVentilatorCumulative | recovered | dataQualityGrade | lastUpdateEt | dateModified | checkTimeEt | death | hospitalized | dateChecked | fips | positiveIncrease | negativeIncrease | total | totalTestResults | totalTestResultsIncrease | posNeg | deathIncrease | hospitalizedIncrease | hash | commercialScore | negativeRegularScore | negativeScore | positiveScore | score | grade | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| date | ||||||||||||||||||||||||||||||||||
| 20200604 | AK | 513.0 | 59584.0 | NaN | 13.0 | NaN | NaN | NaN | 1.0 | NaN | 376.0 | A | 6/4/2020 00:00 | 2020-06-04T00:00:00Z | 06/03 20:00 | 10.0 | NaN | 2020-06-04T00:00:00Z | 2 | 8 | 1907 | 60097 | 60097 | 1915 | 60097 | 0 | 0 | c1046011af7271cbe2e6698526714c6cb5b92748 | 0 | 0 | 0 | 0 | 0 | NaN |
| 20200604 | AL | 19072.0 | 216227.0 | NaN | NaN | 1929.0 | NaN | 601.0 | NaN | 357.0 | 11395.0 | B | 6/4/2020 00:00 | 2020-06-04T00:00:00Z | 06/03 20:00 | 653.0 | 1929.0 | 2020-06-04T00:00:00Z | 1 | 221 | 3484 | 235299 | 235299 | 3705 | 235299 | 0 | 29 | bcbefdb36212ba2b97b5a354f4e45bf16648ee23 | 0 | 0 | 0 | 0 | 0 | NaN |
| 20200604 | AR | 8067.0 | 134413.0 | NaN | 138.0 | 757.0 | NaN | NaN | 30.0 | 127.0 | 5717.0 | A | 6/4/2020 00:00 | 2020-06-04T00:00:00Z | 06/03 20:00 | 142.0 | 757.0 | 2020-06-04T00:00:00Z | 5 | 0 | 0 | 142480 | 142480 | 0 | 142480 | 0 | 26 | acd3a4fbbc3dbb32138725f91e3261d683e7052a | 0 | 0 | 0 | 0 | 0 | NaN |
| 20200604 | AS | 0.0 | 174.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | C | 6/1/2020 00:00 | 2020-06-01T00:00:00Z | 05/31 20:00 | 0.0 | NaN | 2020-06-01T00:00:00Z | 60 | 0 | 0 | 174 | 174 | 0 | 174 | 0 | 0 | 8bbc72fa42781e0549e2e4f9f4c3e7cbef14ab32 | 0 | 0 | 0 | 0 | 0 | NaN |
| 20200604 | AZ | 22753.0 | 227002.0 | NaN | 1079.0 | 3195.0 | 375.0 | NaN | 223.0 | NaN | 5172.0 | A+ | 6/4/2020 00:00 | 2020-06-04T00:00:00Z | 06/03 20:00 | 996.0 | 3195.0 | 2020-06-04T00:00:00Z | 4 | 520 | 4710 | 249755 | 249755 | 5230 | 249755 | 15 | 66 | 1fa237b8204cd23701577aef6338d339daa4452e | 0 | 0 | 0 | 0 | 0 | NaN |
7. Create a module
Finally, you can create a module (eg processing.py) and keep all the above functions in the module. You can simply import them here and use them directly. It will clean the notebook further.
processing.py
While loading the modules, load the “autoreload” extension so that you can change code in the modules and the changes get updated automatically. For more info, see autoreload documentation
%load_ext autoreload
%autoreload 2
from processing import *
The autoreload extension is already loaded. To reload it, use:
%reload_ext autoreload
Advantages
1. Effective for the long run (Maintainability)
Although, the approach may look like an inefficient method of coding but it is very effective in the long run since you will not have to spend hours maintaining the notebook. Given the functions are well written and well defined, they are ready for production.
The code is easily sharable as well as anyone can understand the code unlike in the previous approach. Also, for complex analysis tasks, this approach can be easily used for maintaining the notebook.
2. Proper flow and planning
You do not need to think about the logic of the analysis at the beginning. You can just plan your tasks and write down the required functions which already gives you kind of a framework of mind which helps to be on track. The calm that will follow is likely going to have a greater impact on innovation.
Then, you can finally define the logic at the end to make it work.
3. Easier to modify
You might have noticed that the pipe functionality gives you the ability to modify the tasks or flow easily. You can do so by commenting or adding the functions in the pipeline.
For example, you don’t want to remove the columns and sample the data daily. Then you can achieve this simply by commenting those lines as shown below:
df_daily = (df.pipe(start_pipeline)
.pipe(create_dateindex)
# .pipe(remove_columns)
.pipe(fill_missing)
.pipe(add_state_name)
.pipe(drop_state)
# .pipe(sample_daily)
.pipe(add_active_cases))
4. Easier to debug
In this approach, you know what is happening in each step which makes it a lot easier to debug. Furthermore, since all the operations are functions, you can easily debug the code by performing unit tests or using other methods on the functions.
5. Readability
This approach helps you prevent miles of scrolling and also is easily readable than the previous approach. By looking at the code, you can easily understand what operations are being performed on the data and also can see the effect of those operations on the data in each step using decorator.
Example:
Let us consider cooking chicken. When we do so, we don’t describe the steps like this:
temperature = 210 celsius
food1 = Chicken
food2 = Season(food1, with Spices)
food3 = Season(food2, with Gravy)
Serve(PutInOven(food3, temperature), on a plate)
But instead, we describe it the following way:
temperature = 210 celsius
Chicken.Season(with Spices)
.Season(with Gravy)
.PutInOven(temperature)
.Serve()
The pipe functionality helps us to write code in the latter way, which is also much more readable.
6. Reusability
During production, we turn the project into a Python package. You can import your code and use it in notebooks with a cell. You do not need to write code to do the same task in multiple notebooks.
7. Separation into analysis and data manipulation
Once your functions have been moved to a separate module, two levels of abstraction are obtained: analysis and data manipulation.
You can fiddle around on a high level and keep the details on a low level. The notebook then becomes the summary and a user interface where you can very quickly make nice little charts instead of manipulating data or performing analytical steps to get a result.
Final notes
Hence, following these practices while coding in pandas or performing other similar tasks like building scikit-learn pipelines or other ML pipelines, can be extremely beneficial for developers. Also, all the 4 problems mentioned in the beginning have been solved in this approach. Thus, giving utmost priority to clarity and interoperability, we should remember that it’s a lot easier to solve a problem if we understand the problem well.
Moreover, if you find writing these codes difficult, an open source package called Scikit-lego maintained by Vincent and MatthijsB, with contributions from all around the world, is available. This package does all the hard work for you to create such pipelines along with additional features like custom logging. Do check it out.
Also, if you have any confusion or suggestions, feel free to comment. I am all ears. Thank you.
